Universal-Flex qPCR-Master mix SYBRGreen Blue (2×) and novel ROX-alike reference dye
The qPCR Master mix offers quantitative PCR with a new ROX-alike reference dye for using with all common PCR-Platforms.
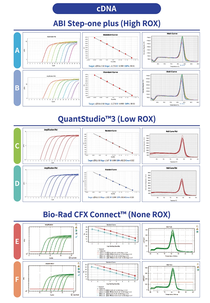
The Universal-Flex qPCR Mastermix is compatible across a variety of instrument platforms including those that require a high, low or no ROX reference signal
ready to use 2x PCR Master mix with SybrGreen and Blue dye. 2 x 1 ml
Components and Mixture
2X BIO-Star Universal-Flex qPCR-Mastermix SYBR Blue contains (2x1 ml):
The master mix is a ready-to-use solution including Hot Start Taq DNA polymerase, dNTPs (dATP, dCTP, dGTP, dTTP) in an optimized PCR buffer system, SybrGreen I, novel passive ROX alike as reference dye and blue inert-dye which does not interfere the PCR-reaction in a real-time Cycler.
Features:
- A novel passive ROX alike reference dye. BIO-Star Universal-Flex
Mastermix is compatible with all common PCR-Platforms
(no ROX, low ROX of high ROX, see Combability list)
- Enzyme with hot start capability increases reaction specificity and sensitivity
- Hot Start Taq DNA polymerase with monoclonal antibodies
- High selectivity and reaction yield
- The mix is featured by a non-fluorescent blue dye for easy pipetting and visual control
Applications:
- Real-time PCR with intercalating dye SYBR Green I
(valid for all ROX concentrations)
- Conventional PCR (cDNA, plasmid DNA, Lambda DNA)
- High-throughput PCR
Description:
The Mastermix contains all components all reagents required for qPCR (except template and primers) The SYBR Green I dye binds to double-stranded DNA formed during real-time PCR. At the same time, the product contains a special ROX Passive Reference Dye, which is suitable for use in all qPCR instruments, and there is no need to adjust the ROX concentration on different instruments.
Taqs: Luna, NEB,
Final price excl. shipping costs3
- verfügbar / avaílable
- 1 - 3 days for delivery / 1 - 3 Tage Lieferzeit1
Storage and
transportation: at -20 °С;. Shipping
with blue ice or at room temperature
Storage
terms: up to 18 months

The Universal-Flex qPCR Mastermix is compatible across a variety of instrument platforms including those that require a high, low or no ROX reference signal
Universal qPCR Mastermix Amplification protocol
1. Defrost the reaction mixture and stir thoroughly.
2. Add the following components into the thin-wall PCR tubes considering the final volume of a reaction mixture equal to 50 μl:
| Component | Volume | Final concentration |
| 2x Mastermix | 25 µl | 1x |
| Forward Primer *1 | variable | 0,2 µM |
| Reverse Primer *1 | variable | 0,2 µM |
| DNA Template *2 | variable | 10 pg - 1 µg |
| Sterile Water | up to 50 µl |
3. Gently vortex and remove droplets by centrifugation.
4. Perform PCR
*1: a: Usually, a good amplification effect can be obtained with the final concentration of 0.2 μM. When the reaction performance is poor, the primer concentration can be adjusted in the range of 0.2-1.0 μM.
*2: b: The amount of template added varies depending on the number of copies of the target gene, and the appropriate amount of template addition is studied by gradient dilution. The best addition amount of template DNA in the 20 µl reaction system was less than 100 ng.
Universal qPCR Mastermix Cycler program
| Step |
Temp. °C |
Incubation time
|
Number of Cycles |
|
Preliminary denaturation |
95 | 0,5-2 min | 1 |
|
Denaturation |
95 | 15 sec | 30-40 |
|
Annealing |
55-65 |
10 sec |
30-40 |
| Elongation | 72 | 30 sec | 30-40 *1 |
|
Melting curve (recommended) |
1 | 1 |
as an alternative:
| Step |
Temp. °C |
Incubation time
|
Number of Cycles |
|
Preliminary denaturation |
95 | 0,5-2 min | 1 |
|
Denaturation |
95 | 15 sec | 30-40 |
|
Annealing / Extension |
60 |
30 sec |
30-40 *1 |
| Elongation | 72 | 30 sec | 30-40 |
|
Melting curve (recommended) |
1 | 1 |
*1: If amplification specificity needs to be improved, two-step procedure or annealing temperature can be used; To improve the amplification efficiency, a three-step procedure or extension time can be used.
Compatible instruments / Cycler List
ABI: 5700, 7000, 7300, 7700, 7900, 7900HT, 7900 HT Fast, StepOne™, StepOne Plus™, 7500/7500 Fast, ViiA 7™,;
Analytik Jena: qTOWER series;
qTOWER: LineGene series
Stratagene: Mx3000P®, 3005P™, 4000™;
Bio-Rad: CFX96™, CFX384™, iCycler iQ™, iQ5™, MyiQ™, MiniOpticon™, Opticon®, Opticon 2, Chromo4™;
Eppendorf: Realplex 2s, Mastercycler® ep, Realplex;
IIIumina: Eco QPCR;
Cepheid: SmartCycler®; QuantStudio™ series, PikoRealTM Cycler
Qiagen Corbett: Rotor-Gene® series;
Roche: LightCycler™ series;
Takara: Thermal Cycler Dice series;
Related products
PCR MASTERMIX SELECTION TABLE
PCR Mastermixes with dTTP and SybrGreen
BioStar 2 PCR mastermix with SYBRGreen, blue dye, no
rox
BioStar 3 PCR mastermix with SYBRGreen, 60 nm LOW-ROX
BioStar 4 PCR mastermix with SYBRGreen, 900 nm HIGH-ROX
PCR Mastermixes with dTTP for probes
BioStar 5 PCR mastermix for probes
BioStar 6 PCR mastermix for probes 60 mM LOW-ROX
BioStar 7 PCR mastermix for probes 900 mM HIGH-ROX
PCR Master Mixes with Evagreen and dUTP
PCR Master Mix for qPCR EVA 1 dUTP+ EvaGreen
PCR Master Mix for qPCR EVA 2 +UNG
PCR Master Mix for qPCR EVA3 +ROX
PCR Master Mix for qPCR EVA4 +ROX+UNG
PCR Master Mix for qPCR EVA5 +low ROX
PCR Master Mix for qPCR E6 +low ROX+UNG
PCR Master Mixes for probes and dUTP
qPCR Master Mix DLP1 Exclusiv +dUTP
qPCR Master Mix DLP2 Exclusiv +dUTP+UDG
qPCR Master Mix DLP3/DLP5 Exclusiv ROX (500 nM, 100nM)
qPCR Master Mix DLP4/DLP6 Exclusiv +ROX 500nM or 100 nM+UDG
Deutsche Beschreibung

Bio-Star Universal-Flex qPCR-Mastermix with SYBRGREEN Blue and a novel ROX-likely reference dye
Bio-Star Universal-Flex qPCR-Mastermix with SYBRGREEN Blue and a novel ROX-likely reference dye
Material Safety Datasheet

References / Protocols / Notes / Recomendations / Tests
Recommendations for avoiding contamination during PCR
Over 10 million copies of DNA template are processed during PCR. Therefore, it is important to prevent the possibility of contamination with other templates and amplicons that are present in laboratory. Here are general recommendations for reducing the risk of contamination:
- Preparation of DNA samples, preparation of reaction solutions, amplification and analysis of PCR products should be carried out in different territorial areas.
- Prepare reaction solutions in PCR laminar flow cabinet equipped with UV lamp.
- Use new pair of gloves when purifying DNA and preparing mixtures and solutions.
- Use reagents designed specifically for PCR. Use pipette tips with integrated aerosol filter when preparing DNA samples and reaction solutions.
- For verification of the absence of contamination, prepare a mixture sample without DNA template (negative control).
Recommendations for primer selection
For design of primers and probe, we recommend using Oligo software http://www.oligo.net/ and its analogs. For selection of oligonucleotides, follow the basic principles:
Primers
- Primer length:18-22 bp.
- Difference in melting temperatures (Тm) of the two primers shouldn't exceed 3 °С.
- Tm of primers for TaqMan PCR should be ≥ 60 °С.
- GC composition of primers should be within the range of 40 to 60%.
- Product length: 70 – 150 bp.
- Minimize secondary structures, avoid them, if possible.
- Check your primers using BLAST.
Probes
- Probe length 22-26 bp;
- Melting temperature: 68-70 °С.
- Minimum of the same nucleotides in a row (especially G: not more than 4 in a row).
- Chose DNA strand that has more C nucleotides than G nucleotides in it.
- There should be no G at 5'-end.
- Avoid self-complementarity and formation of dimers between probe and primers. ( Luna R)
DNA sample
- Purity and integrity of DNA is extremely important for successful PCR. For isolation of DNA, apply conventional methods that allow further amplification of the sample.
- Avoid using PCR inhibitors (phenol, hemin, etc.) when working with the samples. In case of using gel purification, minimize UV exposure in order to prevent formation of pyrimidine dimers.
- Prepare reaction solution in a clean area, use pipette tips with integrated filter in order to reduce contamination risk.
- Optimum amount of DNA per reaction depends on the type of sample and its purity: phage lambda DNA ~0.1 ng; E.Coli DNA ~10 ng; human DNA ~10 – 50 ng.
Characteristics of amplification steps
- Initial DNA denaturation and enzyme activation
- It is very important to achieve complete denaturation of DNA template at the beginning of PCR which provides its efficient use in the first amplification cycle. If GC composition of the
template
is 50% or less, initial denaturation at 95 °С for 5 min will be enough.
Denaturation
Standard time of denaturation per cycle for real-time PCR is considered to be 15 - 30 sec at 95 °C.
Primer annealing and elongation
For TaqMan real-time PCR, annealing and elongation stages are usually combined into one step at 58-60 ºС for 60 sec.
Number of cycles
If there is less than 10 copies of DNA template available per reaction, then efficient amplification requires not less than 40 cycles. A total of 25 – 35 cycles is enough for higher amount of template.