CRISPR - Cas9 Nuclease (S. Pyogenes); CRISPR-associated Cas-System
Clustered Regularly Interspaced Short Palindromic Repeats - "Unlimited" editing of Genes - Accelerator for Molecular Biology studies. Hope Carrier and Breakthrough for the year 2015
Introduction:
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by using CRISPR RNAs (crRNAs) to guide the silencing of invading nucleic acids.
The CRISPR system consists of a short non-coding guide RNA (sgRNA) made up of a target complementary CRISPR RNA (crRNA) and an auxiliary transactivating crRNA (tracrRNA).
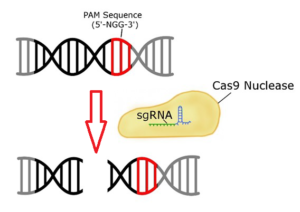
The sgRNA guides the Cas9 endonuclease to a specific genomic locus via base pairing between the crRNA sequence and the target sequence, and cleaves the DNA to create a double-strand break.
The location of the break is within the target sequence 3 bases from the NGG PAM (Protospacer Adjacent Motif). The PAM sequence, NGG, must follow the targeted region on the opposite strand of the DNA with respect to the region complementary sgRNA sequence (Fig.1).
The Cas9 enzyme contains a nuclear localization signal (NLS)
Stability tests / Quality control / Comparison

Cas9 Nuclease functional testing was done by in vitro DNA cleavage assay with the following protocol which gives more than 95% digestion of the substrate DNA as determined by agarose gel electrophoresis.
M: Marker,
UC: Uncut,
1: 1 µl Cas9,
2: 2 µl Cas9
QC-Assay:
Cas9 nuclease is free from detectable RNase, Endonuclease (nicking) and non-specific DNase activities.
References / Protocols / Notes / Recomendations / Tests
1. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. (2012) A programmable dual-RNA-guided, DNA endonuclease in adaptive bacterial immunity. Science.
Aug 17;337(6096):816-21.
2. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. (2013) RNA-guided human genome engineering via Cas9. Science. Feb 15;339 (6121):823-6.
This enzyme is designed to perform CRISPR/Cas9-mediated genome editing
Description
Cas9 Nuclease is the purified recombinant Streptococcus pyogenes Cas9 enzyme containing a nuclear localization signal (NLS) at the C-terminal for targeting to the nucleus. This enzyme is designed to perform CRISPR/Cas9-mediated genome editing (1, 2). The physical purity of this enzyme is ≥98% as assessed by SDS-PAGE with Coomassie® blue staining (Fig. 2).

Recomended Transfection reagents (not provided):
- Nucleofector™ Kits from Lonza
- Lipofectamine™ CRISPRMAX™ from Thermo Fisher
- Electroporation of mammalian cells with Cas9-sgRNA ribonucleoprotein complexes. Any electroporation machine can be used.
Standard Protocol for 30 µl reaction on ice:
|
1.) Target DNA |
x µl - approx 100 ng |
| sgRNA | x µl - approx. 4000 ng |
| 10x Cas9 Reaction Buffer | 3 µl |
| Cas9 Nuclease | 1 µl - approx. 160 ng |
| Water up to | 30 µl |
2) Gently mix the reaction mixture and centrifuge briefly.
3) Incubate at 37 ºC for 60 min.
4) Add 1 µl RNase (4 mg/ml)
5) Incubate at 37 ºC for 20 min.
6) Run 0.7 to1% agarose TBE gel.
Product Source:
E. coli BL21 (DE3) strain expressing a Cas9 gene from Streptococcus pyogenes with an N-terminal 6xHis tag and C-terminal SV40 nuclear localization signal (NLS).
Content:
Cas9 Nuclease in: 50 mM Tris-HCl, 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 50% Glycerol, pH 7.5 @ 25 ºC
Concentration:
Standard-version: 160 ng/µl
HC-Version: 1600 ng/µl
10x Cas9 Nuclease Reaction Buffer (200 mM HEPES, 1000 mM NaCl, 50 mM MgCl2, 1 mM EDTA, pH 6.5 @ 25 °C)
Storage: at -20°C for 24 months, avoid frequent thawing and freezing
Transport: with blue ice
Cas9 Nuclease is the purified recombinant Streptococcus pyogenes Cas9 enzyme containing a nuclear localization signal (NLS) at the C-terminal for targeting to the nucleus. This enzyme is designed to perform CRISPR/Cas9-mediated genome editing (1, 2). The physical purity of this enzyme is ≥98% as assessed by SDS-PAGE with Coomassie® blue staining.
* availibility of sample size may be limited
Final price excl. shipping costs3
- verfügbar / avaílable
- 1 - 3 days for delivery / 1 - 3 Tage Lieferzeit1
Deutsche Beschreibung

Cas 9 Nuklease - CRISPR
Datasheet Cas9 Nuclease for CRISPR Method
Material Safety Datasheet